Coding Genotypes
The field names for marker information have to be constructed very carefully, because the core code uses these names to understand what data is being presented to it. Each field name has a number of sections, separated by underscores.
For example, the field “g_2_3_M1-_1_2” conveys that the field contains genotype marker for a 2-allele marker with 3 genotypes considered (genotypes 11, [12, 21] and 22), the marker name is “M1-” and the allele names are “1” and “2”.
Mappings for Genetic Markers
Section titled “Mappings for Genetic Markers”Use these mappings to construct the marker header name:
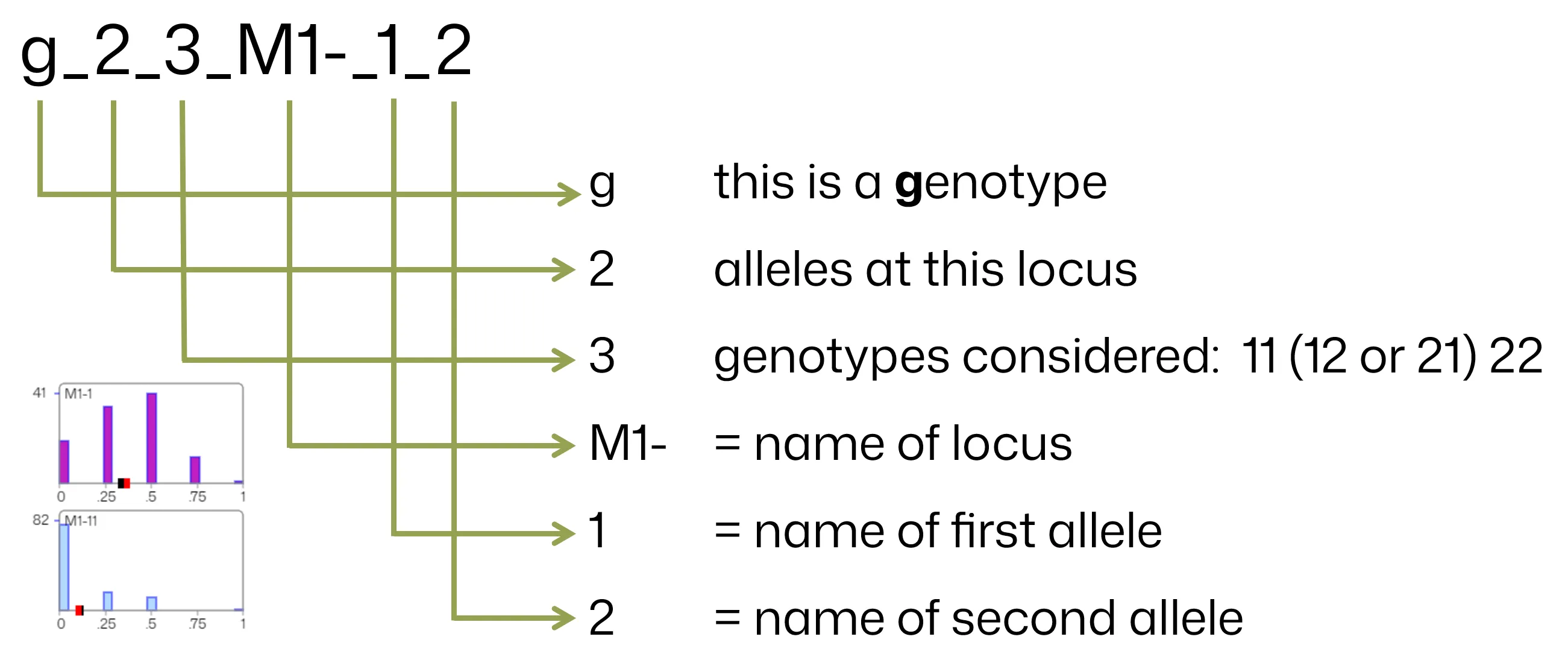
Below is another example for illustration:
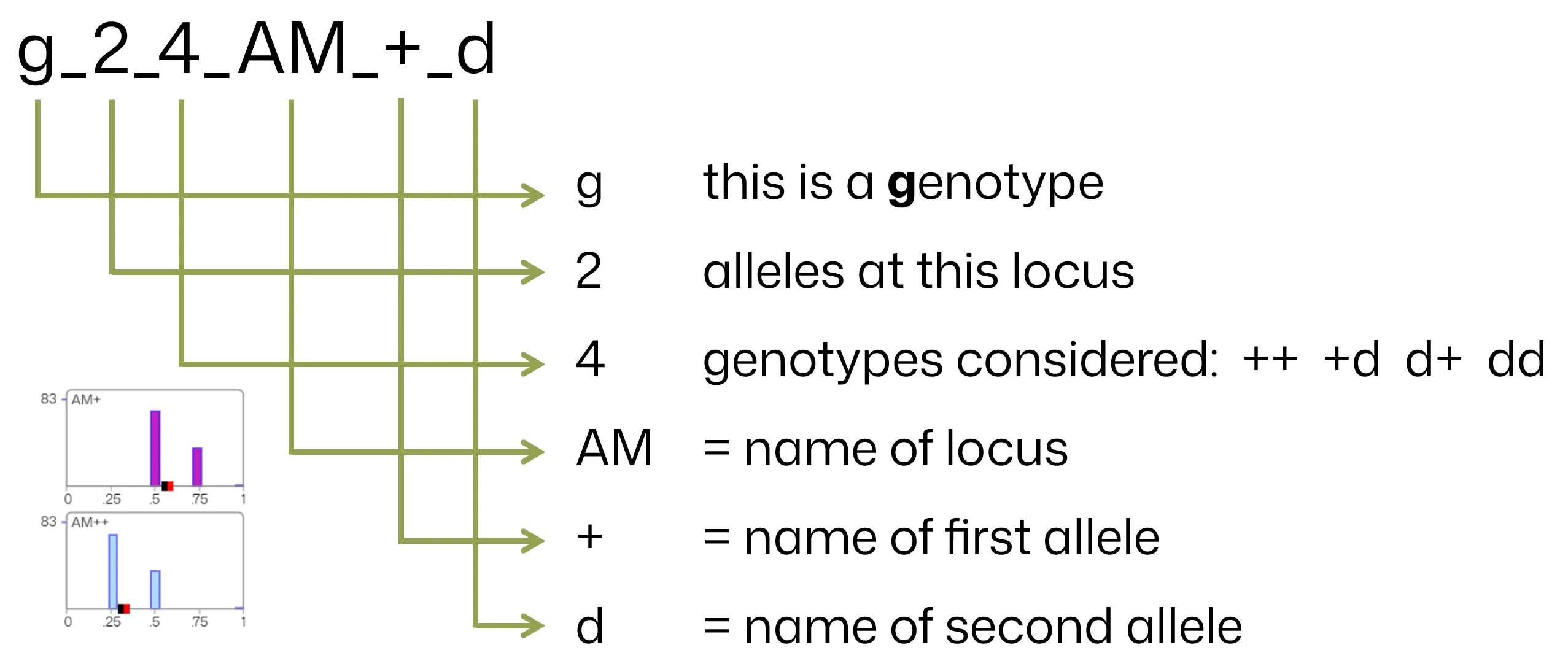
Component Details
Section titled “Component Details”The table below describes further each component:
| Component | Description |
|---|---|
| Type | ”g” or “G” for a genotype. Genotypes have only one field/column per marker. |
| Number of alleles (nA) | Usually 2, but any number > 2 is accepted. |
| Number of genotypes | This is the number of genotypes that could be present, including lethals, even if not all genotypes are in your dataset. So if you have no 22 genotypes in your dataset, and you are grouping 12 and 21 together as one genotype, then these are the genotypes in this list: [11, (12 or 21), 22], and the number of genotypes is 3, not 4. |
| Marker/Locus Name | It is recommended to append a dash to the marker name as in the “M1-” example above because marker name is usually followed directly by the name of one of its alleles in graphical output. Do not use a space in a name. Marker names should not be longer than 12 characters. |
| Allele 1 Name | Alphanumeric text. Do not use a space in a name. No longer than 4 characters. |
| Allele 2 Name | As above |
| Allele n name | As above, name all nA alleles |
Candidate Values
Section titled “Candidate Values”The content above just tells us how to construct the header name for a genetic marker, but what values do we enter for each candidate?
As a simple rule, the range of valid marker values for a candidate will be from 1 to Number of genotypes (inclusive) as below:
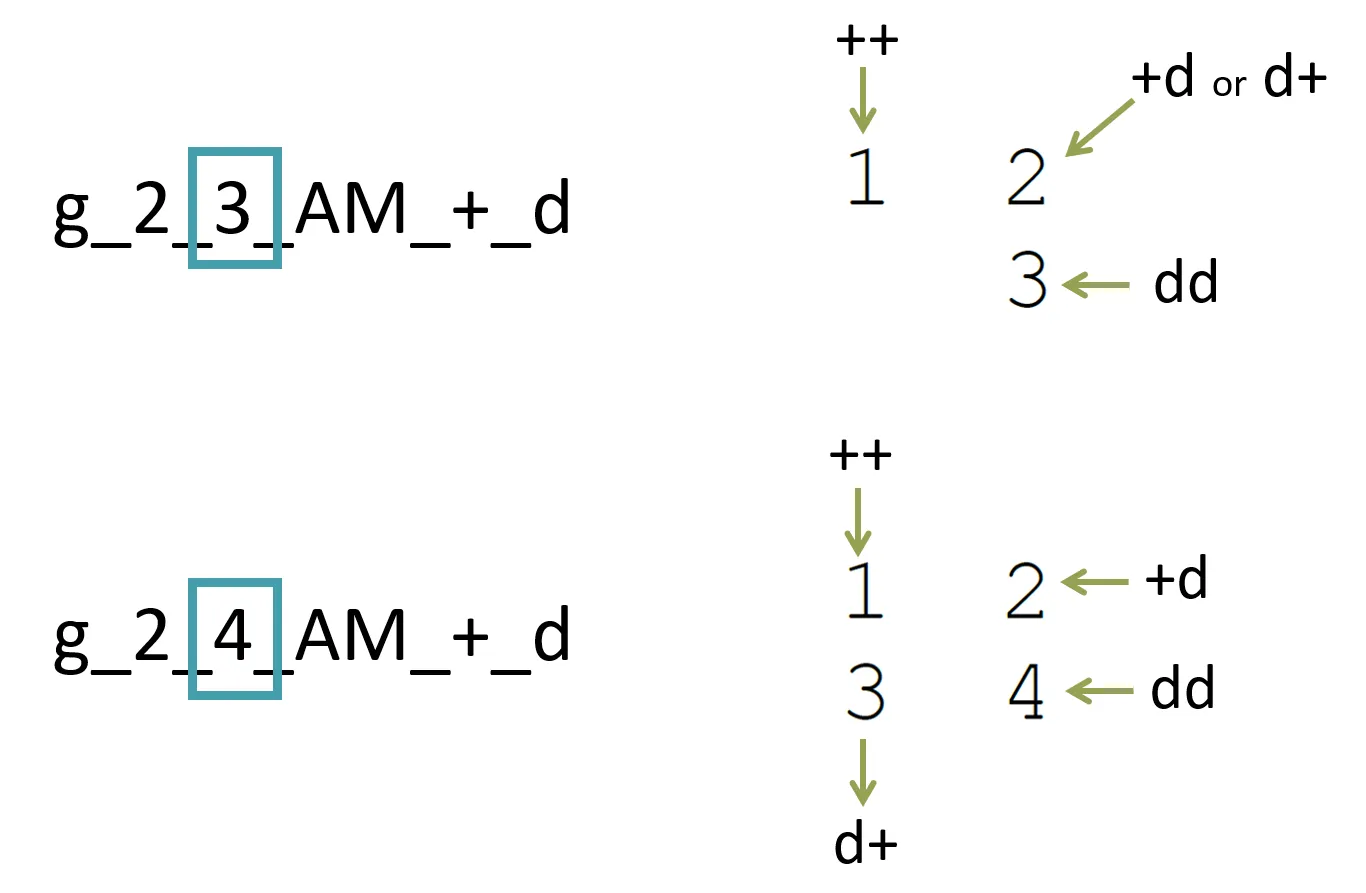
Let’s assume:
- Allele ”+” represents the favourable allele
- Allele “d” represents the defective allele
- Hence the “dd” genotype represents an affected candidate
In the first example above, any known affected candidate would have the value “3”. In the second example above, any known affected candidate would have the value “4”.
Using MateSel, you now have the option to:
- Ensure there are no “dd” genotypes in the progeny.
- Increase the allele frequency of the favourable ”+” allele with each new generation
Missing Genotypes
Section titled “Missing Genotypes”Genotypes denoted as 0 or a period . are treated as missing. An individual with a missing genotype is allocated the raw mean of genotype probabilities for current candidates (i.e. excluding those who are only listed as candidates in CommittedMatings.txt) that have legal genotypes.

