Manage non-additivity: Genomic Mate Selection (GMS)
Normally, MateSel predicts progeny trait merit as the average of the parents EBVs. This works for most traits (and even things treated as traits, like age).
But it’s not correct when offspring are expected to deviate from the parental average because of:
- Dominance effects
- Increased segregation variance
- Breeding costs or compatibility issues
In these cases, we need a way to model the non-additive deviations. As some form of mate selection based on genomic information seems a most likely application, in MateSel we call this facility Genomic Mate Selection (GMS).
GMS Data Files
Section titled “GMS Data Files”Non-additive information can’t go in the main data file (since it only has one line per candidate). Instead, you supply it in separate data files, one per trait. Each trait GMS file contains the predicted non-additive deviation for each possible pairing of a male and a female candidate. Here is how to setup the files:
- Main Data File: Name GMS traits starting with GMS_ (e.g. GMS_BodyWeight). It’s case-sensitive: renaming it (e.g. gMS_BodyWeight) turns GMS off.
- GMS Trait Files: For each GMS trait, create a file:
- candGMS<traitname>.txt where traitname is the exact trait name in your main datafile e.g. candGMS_BodyWeight.txt
- Examples of GMS files
- candGMS_BFdom.txt → genomic dominance deviations for Backfat
- candGMS_BFprogSD.txt → genomically derived predictions of segregation variance for Backfat
- candGMS_PedInb.txt → pedigree-derived inbreeding coefficients (for illustration)
- candGMS_GRMInb.txt → genomically-derived inbreeding coefficients (for illustration)
- candGMS_Must.txt → must-have matings
- candGMS_MustNot.txt → must not have matings
- candGMS_Ind-100F.txt → to make and index of: Index minus 100 times progeny F
Values you can use in each GMS File:
- Enter additive EBVs into main data file, and put only the non-additive part in the candGMS file OR
- Set them all to 0 in the main data file, and put the total progeny merit (additive + non-additive) in the candGMS file.
Example:
candGMS_PedInb Lilianna Lisa Rayna …Samir 0.15493 0.04952 0.06282 …Jeff 0.02658 0.07900 0.02700 …Magnus 0.03528 0.03417 0.03828 …… … … …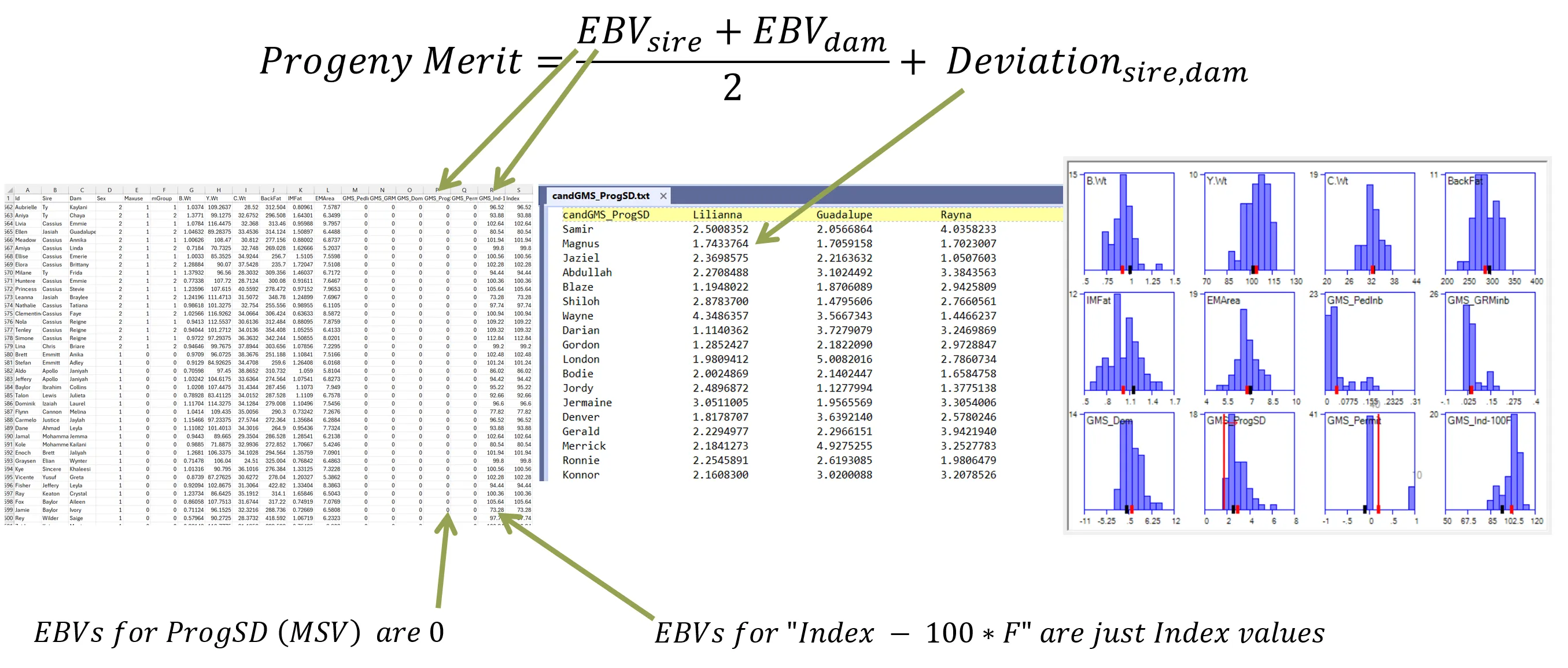
Some rules on the GMS format example above:
- A top-left element (candGMS_PedInb above) is required, but just for the read formatting. It can be any text containing no spaces.
- Order of candidates is not important.
- Surplus candidates that are not in the present run are OK. They are ignored.
- Include candidates that are not candidates in your main datafile, but are entered in CommittedMatings.txt.
- Under bisexuality, for an individual that is available as both a male candidate and a female candidate, it should have its own row as well as its own column. You can control pattern of selfing using, for example, -1 or 1 for the self x self elements.
- If there are missing candidates, this will be reported in the logs.
- For missing element information, you can use the value of -999999. These will be reported in the logs.
Video Demonstration
Section titled “Video Demonstration”The video below further explains how to use GMS. It uses the older “MateSel Legacy” interface, but the feature works the same in newer versions.
GMS Output
Section titled “GMS Output”GMS traits are monitored and controlled using the same histograms as standard additive (non-GMS) traits.
In addition, a file OutGMSnon-add.txt will be written to the same location as your main data file after the run stops. These show the non-additive component for each GMS trait for each mating in the solution. The additive components are listed in other output files, as for normal non-GMS traits.
GMS and other features
Section titled “GMS and other features”The GMS feature operates simultaneously with:
- Grouping
- Multiple EndUses
- Mixed Mating Groups
- Committed Matings
Examples
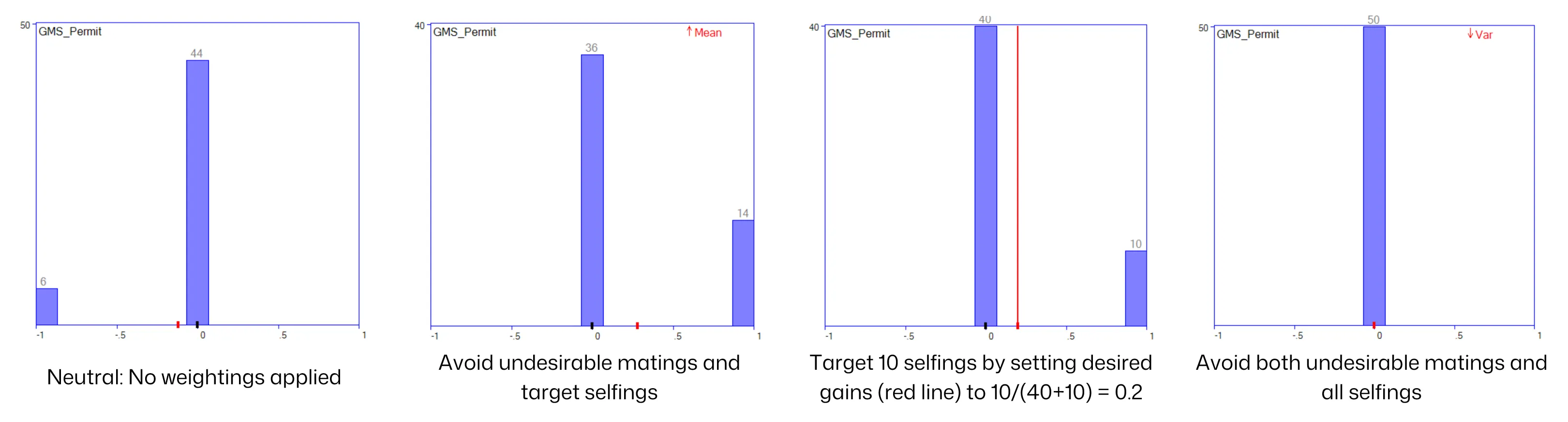
Section titled “Examples”Managing undesirable (-1) and desirable (1) matings: Just for illustration, assume that desirable matings are user-nominated selfings, and that you want to maximise or otherwise control the number of these. Here are some outcomes:
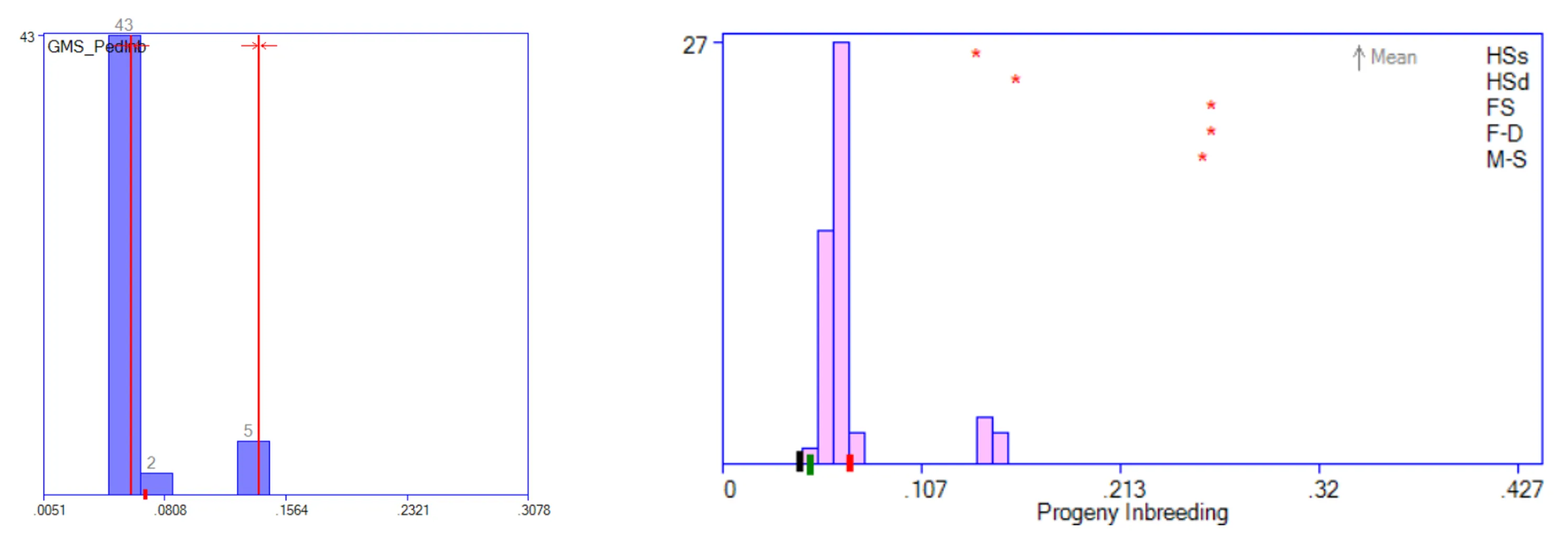
Progeny Inbreeding: This is a simple test of GMS operation. For trait GMS_PedInb, bimodality was targeted, with 10 percent of matings close to F=0.14 and the remainder close to F=0.06. The results are the same as for the Progeny Inbreeding histogram itself:
Potential Applications
Section titled “Potential Applications”The GMS facility could be used to accommodate:
- Phenotypic effects on mating/pregnancy/birth performance (e.g. A small male might not reach a big female, or a big bull on a small cow could lead to dystocia).
- Animal behaviour compatibility scores for each possible male x female mating.
- The predicted transport cost associated with making each possible male x female mating.
- Imprinting effects, whereby some aspect of genetic merit depends on the gender of each parent involved.
- Avoiding the duplication of matings that were already made in a previous mating round.
- Make a new GMS trait with zero values in the main data file.
- Set all non-additive values to 0, except for the male x female elements that represent matings to be avoided – set them to 1.
- Use the trait management tools to ensure there are no progeny with a value of 2 (1+1)
GMS in Research and Industry
Section titled “GMS in Research and Industry”There is an increasing interest is this area, with frequent new academic publications on how to use genomic information for increasing both short-term and longer-term genetic gains. Total Genetic Merit in progeny: Refined predictions of progeny merit can involve dominance components, and possibly some epistatic components, at nominated QTL. It could also involve genome-wide heterozygosity, although that may be less powerful. The Expected Cross Value of Ahadi et al. (https://www.nature.com/articles/s41437-024-00697-y) goes beyond this to consider the value of a mating pair as a function of allelic information and transmission probabilities for each partner, increasing the desirable allele frequency.
Look Ahead Mating Schemes: “Genomic Mating uses information in a similar fashion to genomic selection but includes information on complementation of parents to be mated” (Akdemir and Sanchez, 2016. https://doi.org/10.3389/fgene.2016.00210).
With thanks to Tobias Niehoff for clarifying and expanding on our earlier effort surrounding his work:
The Usefulness criterion (Schnell & Utz, 1975; Lehermeier et al. https://doi.org/10.1534/genetics.117.300403) describes the expected genetic level of selected progeny of a cross and has received increasing interesting in recent years as estimated SNP effects allow the prediction of progeny variance. Important for use with the GMS facility is that the usefulness criterion is expressed for a specific mating pair of inbred lines or individuals. Criteria that describe a comparable property as the usefulness criterion and utilize segregating variance in gametes have been proposed for animal breeding (e.g. Bijma et al., 2020, https://doi.org/10.1534/genetics.119.302643). Criteria Index5 and Index6 of Bijma et al. avoid the combinatorial issue that a property of offspring of a mating is expressed for a mating pair by assuming that the gametic Mendelian Sampling variance of the mating partner is about the same as for the focal individual (i.e., assuming the heterozygosity level and linkage of the unknown mating partner are comparable). Thus, these criteria may be entered as if they are EBVs in the MateSel main datafile for maximization or other manipulation. Methods ProbSelOff and ExpBVSelOff of Niehoff et al. (2024) (https://doi.org/10.1186/s12711-024-00899-2) are the equivalent versions of Index5 and Index6 which do not make assumptions about the mating partner and would have to be provided in the candGMS_XXX.txt file. Niehoff et al. (2024) also extended the planning horizon by one more generation to the grand-offspring generation with their criterion ProbSelGrOff and ExpBVSelGrOff which not only consider the segregation variance of gametes of selected parents but also of their progeny, and thus aim for increased genetic variation grand-progeny, and thus increased genetic merit in great-grandprogeny. These criteria are also expressed for a particular mating pair, meaning they also have to be supplied through the candGMS_XXX.txt file. Schnell, F., & Utz, H. (1975). Bericht über die Arbeitstagung der Vereinigung österreichischer Pflanzenzüchter. Gumpenstein, Austria: BAL Gumpenstein, 243-248.
Moeinizade et al. (https://doi.org/10.1534/g3.118.200842) aimed a number of generations ahead using an approximate objective function of genetic merit in the last generation, involving simulation. However, for this method, “The contribution of a breeding parent is evaluated based on not only the favorable alleles that it carries but also the favorable alleles that it carries but are missing in other selected breeding parents.” This means that the elements of the GMS matrix are dynamic, and cannot be read-in up-front. For such cases, further development of MateSel would be required given ‘on-the-fly’ calculation of the required elements, or indeed merit across matings, in light of the current mating set, including Committed Matings. Alternatively, direct calling of external routines that calculate such values could be developed in collaboration with others. Let us know if you have an interest in doing this.

